PPOL 5203 Data Science I: Foundations
Tidy Data and Joining Methods in Pandas
Tiago Ventura
Tidy Data and Joining Methods in Pandas
Tiago Ventura
In this Notebook we cover
This is our last notebook of data wrangling with Pandas. We will cover:
- Tidy Data
- Joining Methods in Pandas
import pandas as pd
import numpy as np
Tidy Data¶
Data can be organized in many different ways and can target many different concepts. Having an consistent and well-established procedure for data organization will make your workflow, faster, more efficient and less prone to errors.
On tasks related to data visualization and data wrangling, we will often try to organize our datasets following a tidy data format proposed by Hadley Wickham in his 2014 article.
Consider the following 4 ways to organize the same data (example pulled from R4DS).
| Country | Year | Cases | Population |
|---|---|---|---|
| Afghanistan | 1999 | 745 | 19987071 |
| Afghanistan | 2000 | 2666 | 20595360 |
| Brazil | 1999 | 37737 | 172006362 |
| Brazil | 2000 | 80488 | 174504898 |
| China | 1999 | 212258 | 1272915272 |
| China | 2000 | 213766 | 1280428583 |
| country | year | type | count |
|---|---|---|---|
| Afghanistan | 1999 | cases | 745 |
| Afghanistan | 1999 | population | 19987071 |
| Afghanistan | 2000 | cases | 2666 |
| Afghanistan | 2000 | population | 20595360 |
| Brazil | 1999 | cases | 37737 |
| Brazil | 1999 | population | 172006362 |
| country | year | rate |
|---|---|---|
| Afghanistan | 1999 | 745/19987071 |
| Afghanistan | 2000 | 2666/20595360 |
| Brazil | 1999 | 37737/172006362 |
| Brazil | 2000 | 80488/174504898 |
| China | 1999 | 212258/1272915272 |
| China | 2000 | 213766/1280428583 |
| country | 1999 |
2000 |
|---|---|---|
| Afghanistan | 745 | 2666 |
| Brazil | 37737 | 80488 |
| China | 212258 | 213766 |
| country | 1999 |
2000 |
|---|---|---|
| Afghanistan | 19987071 | 20595360 |
| Brazil | 172006362 | 174504898 |
| China | 1272915272 | 1280428583 |
What makes a data tidy?¶
Three interrelated rules which make a dataset tidy:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.

Why Tidy?¶
There are many reasons why tidy format facilitates data analysis:
- Facilitates split-apply-combine analysis
- Take full advantage of
pandasvectorize operations over columns, - Allows for apply operations over units of your data
- Sits well with grammar of graphs approach for visualization
Using pandas to tidy your data.¶
Most often you will encounter untidy datasets. A huge portion of your time as a data scientist will consist on apply tidy procedures to your dataset before starting any analysis or modeling. Let's learn some pandas methods for it!
Let's first create every example of datasets we saw above
base_url = "https://github.com/byuidatascience/data4python4ds/raw/master/data-raw/"
table1 = pd.read_csv("{}table1/table1.csv".format(base_url))
table2 = pd.read_csv("{}table2/table2.csv".format(base_url))
table3 = pd.read_csv("{}table3/table3.csv".format(base_url))
table4a = pd.read_csv("{}table4a/table4a.csv".format(base_url), names=["country", "cases_1999", "cases_2000"] )
table4b = pd.read_csv("{}table4b/table4b.csv".format(base_url), names=["country", "population_1999", "poulation_2000"] )
table5 = pd.read_csv("{}table5/table5.csv".format(base_url), dtype = 'object')
# see
table1
For most real analyses, you will need to resolve one of three common problems to tidy your data:
One variable might be spread across multiple columns (
pd.melt())One observation might be scattered across multiple rows (
pd.pivot_table())A cell might contain weird/non-sensical/missing values.
We will focus on the first two cases, as the last requires a mix of data cleaning skills that are spread over different notebooks
pd.melt() from wide to long¶
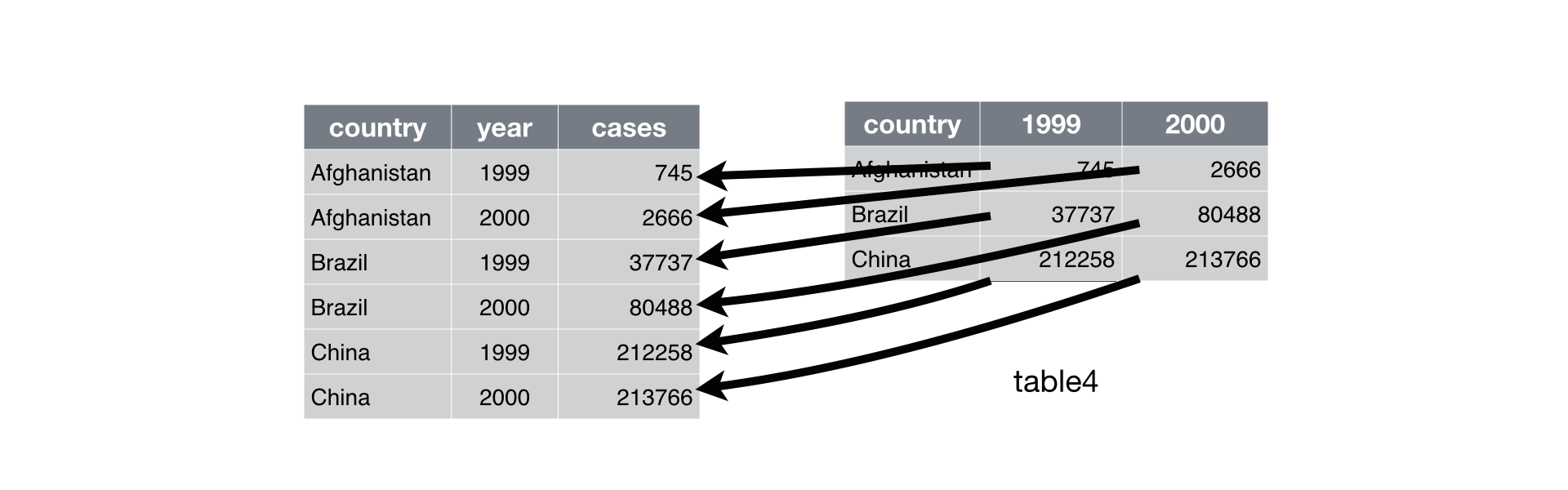
Requires:
Columns whose names are identifier variables, and you wish to keep in the dataset as it is.
A string for the new column with the variable names.
A string for the new column with the values.
# untidy - wide
print(table4a)
# tidy - from wide to long
table4a.melt(id_vars=['country'], var_name = "year", value_name = "cases")
help(pd.melt)
pd.pivot_table() from long to wide (but tidy)¶
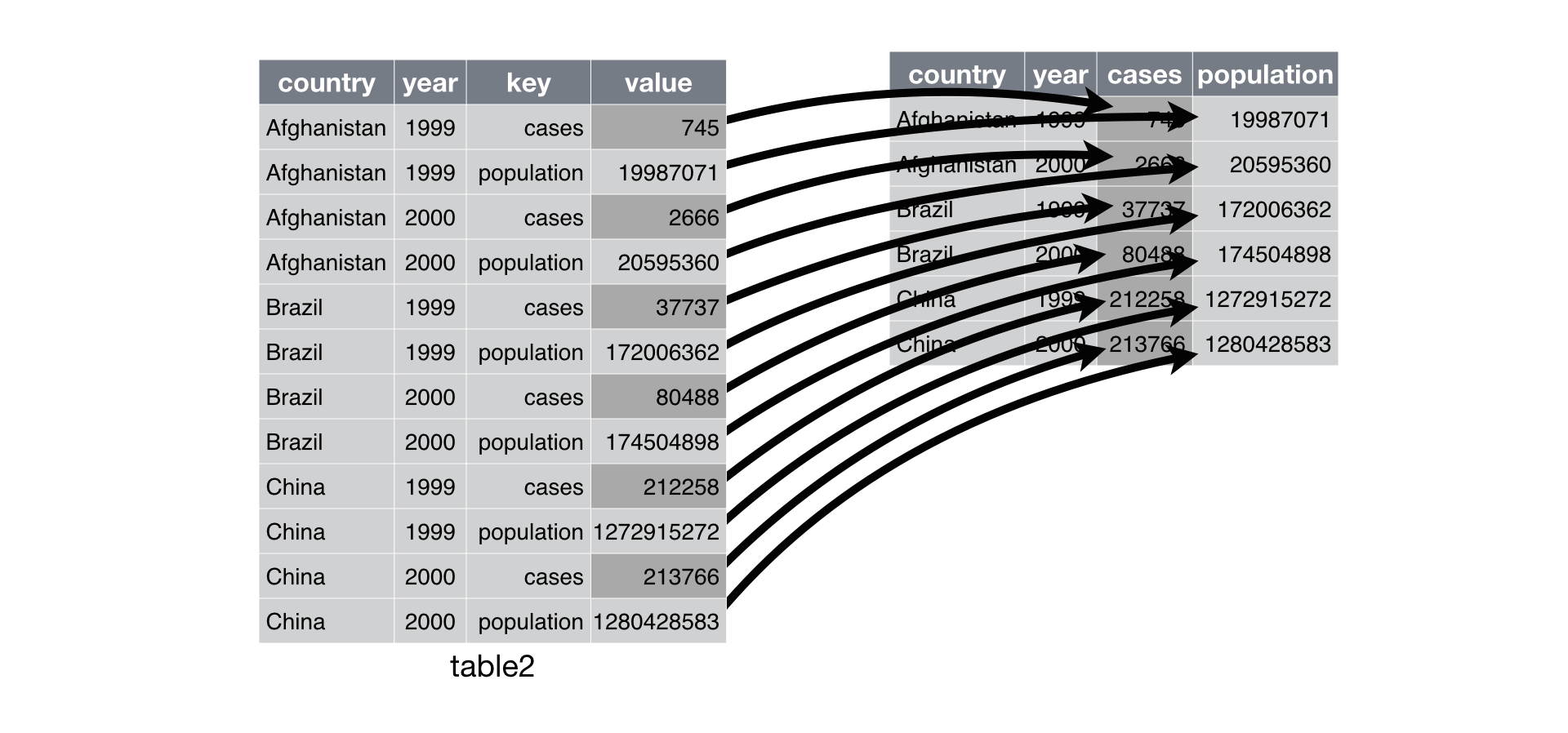
pivot_table() is the opposite of melt(). Think about this as you are widening a coarced variable. It requires:
Index to hold your new dataset upon
The column to open up across multiple new columnes.
The column with the values to fill the cell on the new colum.
#untidy
print(table2)
#tidy
table2_tidy = table2.pivot_table(
index = ['country', 'year'],
columns = 'type',
values = 'count')
# see
table2_tidy.reset_index()
help(pd.pivot_table)
string methods with pandas¶
To tidy example three, we will use a combination of pandas and string methods. This combination will be around very often in our data cleaning tasks
# we need to separate rate
table3
str.split(): to split strings in multiple elements¶
# simple string
hello, world = "hello/world".split("/")
print(hello)
print(world)
# with pandas
table3[["cases","population"]]=table3['rate'].str.split("/", expand=True)
table3
string methods will be super helpful on cleaning your data
# all to upper
table3["country_upper"]=table3["country"].str.upper()
table3
# length
table3["country_length"]=table3["country"].str.len()
# find
table3["country_brazil_"]=table3["country"].str.find("Brazil")
# replace
table3["country_brazil"]=table3["country"].str.replace("Brazil", "BR")
# extract
table3[["century", "years"]] = table3["year"].astype(str).str.extract("(\d{2})(\d{2})")
# see all
table3
Joining Methods¶
It is unlikely that your work as a data scientist will be restricted to analyze one isolated data frame -- or table in the SQL/Database lingo. Most often you have multiple tables of data, and your work will consist of combining them to answer the questions that you’re interested in.
There two major reasons for why complex datasets are often stored across multiple tables:
A) Integrity and efficiency issues often referred as database normalization. As your data grow in size and complexity, keeping a unified database leads to redundancy and possible errors on data entry.
B) Data comes from different sources. As a researcher, you are being creative and augmenting the information at your hand to answer a policy question.
Database normalization works as an **constraint**, a guardrail to protect your data infrastructure. The second reason for why joining methods matter is primarily an **opportunity** . Keep always your eyes open for creative ways to connect data sources. Very critical research ideas might emerge from data augmentation from joining unrelated datasets.
pandas methods:¶
pandas comes baked in with a fully functional method (pd.merge) to join data. However, we'll use the SQL language when talking about joins to stay consistent with SQL and R Tidyverse.
Let's start creating two tables for us to play around with pandas join methods
# Two fake data frames
import pandas as pd
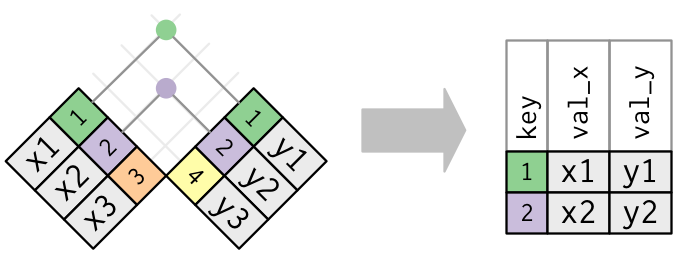
data_x = pd.DataFrame(dict(key = ["1","2","3"],
var_x = ["x1","x2","x3"]))
data_y = pd.DataFrame(dict(key = ["1","2","4"],
var_y = ["y1","y2","y4"]))
display(data_x)
display(data_y)
# chaining datasets
data_x.merge(data_y,how="left")
# calling the construct
pd.merge(data_x, data_y,how="left")
# chaining datasets
data_x.merge(data_y,how="right")
# chaining datasets
data_x.merge(data_y,how="outer")
# chaining datasets
data_x.merge(data_y,how="inner")
Handling disparate column names¶
# rename datasets
data_X = data_x.rename(columns={"key":"country_x"})
data_Y = data_y.rename(columns={"key":"country_y"})
# join now, and you will get an error
pd.merge(data_X,
data_Y,
how="left",
left_on = "country_x", # The left column naming convention
right_on="country_y") # The right column naming convention )
help(pd.merge)
Concatenating by columns and rows¶
By Rows: pd.concat(<>, axis=0)¶

# full of NAS because the second columnes do not have the same name
pd.concat([data_x,data_y],
sort=False) # keep the original structure
By Columns: pd.concat(<>, axis=1)¶

pd.concat([data_x,data_y],axis=1)
Note that when we rowbind two DataFrame objects, pandas will preserve the indices. And you can use this to sort your dataset.
pd.concat([data_x,data_y],axis=0).sort_index()
To keep the data tidy, we can preserve which data is coming from where by generating a hierarchical index using the key argument. Of course, this could also be done by creating a unique columns in each dataset before the join.
pd.concat([data_x,data_y],axis=0, keys=["data_x","data_y"]).reset_index()
Lastly, note that we can completely ignore the index if need be.
pd.concat([data_x,data_y],axis=0,ignore_index=True)
Practice¶
Using both the World Cup Datasets from the Data Wrangling Notebook:
Select the following columns: MatchID, Year, Stage, Home Team Name, Away Team Name, Home Team Goals, Away Team Goals.
Clean the column names converting all to lower case and removing spaces.
Convert this data to the long format, having: a) a single column for the teams playing; b) a single column for the goals scored.
Which team have the higher average of goals in the history of the world cup?
# open datasets
wc = pd.read_csv("WorldCups.csv")
wc_matches = pd.read_csv("WorldCupMatches.csv")
# 1 and 2
wc_copy = (wc_matches.filter(["MatchID", "Year", "Stage",
"Home Team Name",
"Away Team Name", "Home Team Goals",
"Away Team Goals"]).
rename(columns={col: col.lower().replace(" ", "_") for col in wc_matches.columns}).copy()
)
wc_copy
# rotating the team names
wc_home = (wc_copy.
filter(["matchid", "year", "stage", "home_team_name", "away_team_name"]).
melt(id_vars=["matchid", "year", "stage"],
var_name="team_host",
value_name="team_name").
assign(team_host= lambda x: x["team_host"].str.replace("_team_name", ""))
)
wc_home
# 3
wc_goals = (wc_copy.
filter(["matchid", "year", "stage", "home_team_goals", "away_team_goals"]).
melt(id_vars=["matchid","year", "stage"],
var_name="team_host",
value_name="team_goals").
filter(["matchid","team_host", "team_goals"])
)
wc_goals
wc_home.head()
wc_goals.head()
wc_tidy = pd.concat([wc_home, wc_goals],axis=1)
# 4
(wc_tidy
.groupby(["team_name"], as_index=False)
["team_name"]
.size()
.sort_values("size", ascending=False)
)
# 5
(wc_tidy
.groupby(["team_name"], as_index=False)
["team_goals"]
.agg(["mean", "count"])
.sort_values("mean", ascending=False).
head(10)
)
!jupyter nbconvert _week-5c-joining_data.ipynb --to html --template classic