PPOL 5203 Data Science I: Foundations
Data Wrangling in Pandas
Tiago Ventura
Data Wrangling in Pandas
Tiago Ventura
In this Notebook:
In this notebook, we will cover standard data wrangling methods using pandas:
- Selecting Methods.
- Filtering Methods.
- Grouping and Summarization.
- Recoding Variables.
Setup¶
import pandas as pd
import numpy as np
# set some options for pandas
pd.set_option('display.max_rows', 10)
Data Wrangling in pandas¶
Since you are also learning R throughout DSPP, let's provide you with an overview of the main Data Wrangling Functions in R using Tidyverse and Python using pandas.
pandas |
dplyr$^\dagger$ |
Description |
|---|---|---|
.filter() |
select() |
select column variables/index |
.drop() |
select() |
drop selected column variables/index |
.rename() |
rename() |
rename column variables/index |
.query() |
filter() |
row-wise subset of a data frame by a values of a column variable/index |
.assign() |
mutate() |
Create a new variable on the existing data frame |
.sort_values() |
arrange() |
Arrange all data values along a specified (set of) column variable(s)/indices |
.groupby() |
group_by() |
Index data frame by specific (set of) column variable(s)/index value(s) |
.agg() |
summarize() |
aggregate data by specific function rules |
.pivot_table() |
spread() |
cast the data from a "long" to a "wide" format |
pd.melt() |
gather() |
cast the data from a "wide" to a "long" format |
.() |
%>% |
piping, fluid programming, or the passing one function output to the next |
If you want to fully embrace the tidyverse style from R in Python, you should check the dfply module. This modules ofers an alternative to data wrangling in Python, and mirrors the popular tidyverse functionalities from R.
We will not cover dfply in class because I believe you should dominate pandas as data scientists that are fluent in Python. However, feel free to learn and even use in your homeworks and assignment.
# load worldcup datasets
wc = pd.read_csv("WorldCups.csv")
wc_matches = pd.read_csv("WorldCupMatches.csv")
# see wc data
wc.head()
# see matched data
wc_matches.head()
Piping¶
If you started your data science career, or are simultaneously learning R, it is likely you were exposed to the tidyverse world, and the famous pipe. Here it is for you:

Pipe operators, introduced by tha magritt package in R, absolutely transformed the way of writing coding in R. It did for good reasons. And in the past few years, piping has also become more prevalent in Python.
Pipe operators allows you to:
- Chain together data manipulations in a single operational sequence.
In pandas, piping allows you to chain together in a single workflow a sequence of methods. Pipe operators (.pipe()) in pandas were introduced relatively recently (version 0.16.2).
Let's see some examples using the world cup data. We want to count which country played more world cup games.
# read world cup data
wc = pd.read_csv("WorldCupMatches.csv")
Method 1: sequentially overwrite the object¶
wc = pd.read_csv("WorldCupMatches.csv")
# select columns
wc = wc.filter(['Year','Home Team Name','Away Team Name'])
# make it tidy
wc = wc.melt(id_vars=["Year"], var_name="var", value_name="country")
# group by
wc = wc.groupby("country")
# count occurrences
wc = wc.size()
# move index to column with new name
wc = wc.reset_index(name="n")
# sort
wc = wc.sort_values(by="n", ascending=False)
# print 10
wc.head(10)
Method 2: Pandas Pipe¶
wc = pd.read_csv("WorldCupMatches.csv")
# select columns
wc_10 = (# encapsulate
wc.filter(['Year','Home Team Name','Away Team Name']).
melt(id_vars=["Year"], var_name="var", value_name="country").
groupby("country").
size().
reset_index(name="n").
sort_values(by="n", ascending=False)
)# close
# print 10
wc_10.head(10)
Final notes in Piping¶
It should be clear by now, but these are some of the advantages of piping your code:
- Improves code structure
- Eliminates intermediate variables
- Can improve code readability
- Memory efficiency by eliminating intermediate variables
- Makes easier to add steps anywhere in the sequence of operations
Keep in mind pipes also have some disadvantages. In my view, specially when working in notebook in which you do not execute line by line, pipes can make codes a bit harder to debug and also embed errors that could be more easily perceived by examining intermediate variables.
Select Columns¶
Functionality:
- Select specific variables/column indices
Implementation
- Traditional indexing in pandas
.loc()methodspd.filter()methods (allows piping).
Select via index¶
# load worldcup dataset
wc = pd.read_csv("WorldCups.csv")
wc_matches = pd.read_csv("WorldCupMatches.csv")
## simple index for columns
wc["Year"]
## using dot method
wc.Year
## using .loc method
wc.loc[:,"Year"]
Notice, in all cases, the output comes as a pandas series. If you would like the output to be a full data frame, or if you need to select multiple columns, you should give a list of indexes as inputs
wc[["Winner","Year"]]
.loc() methods for selecting columns¶
It allows you to select multible variables in between column names!
# .loc for a single or multiple columns
wc.loc[: , "Year":"Winner"]
# iloc for numeric positions
wc.iloc[0:10,0:3]
.filter() method¶
The filter methods in Pandas works similarly to the select function in the Tidyverse in R. It has the following advantages:
- Allows for a piping approach
- Can be combined with regex queries for selectins columns
# simple filter
wc.filter(["Year", "Winner"])
Using the like parameter: Select columns that contain a specific substring.
# like parameter. In R: data %>% select(contains("Away"))
wc_matches.filter(like="Away")
Using the Regex: You can also input regex queries for selecting columns. More and More flexibility
# starts with
wc_matches.filter(regex="^Away")
# ends with
wc_matches.filter(regex="Initials$")
Drop Columns¶
Functionality:
Drop specific variables/column indices
Implementation:
- Indexing + Boolean operations
- .loc() methods
- pd.drop() methods (allows piping).
## select all but "Year" and "Country"
wc[wc.columns[~wc.columns.isin(["Year", "Country"])]]
# Indexing: Bit too much?!
# .isin returns a boolean.
wc.columns[~wc.columns.isin(["Year", "Country"])]
# What is going on here?
~wc.columns.isin(["Year", "Country"])
# loc methods
wc.loc[:,~wc.columns.isin(["Year", "Country"])]
# indexing -> error
# it indexes boolean with rows.
bol_col = ~wc.columns.isin(["Year", "Country"])
bol_col
wc[bol_col]
# With .loc, it works
wc.loc[:,bol_col]
pd.drop() methods: easier way to go¶
wc.drop(columns=["Year"])
# see here why you need the argument columns
help(pd.DataFrame.drop)
Create new columns¶
Functionality:
- Create a new column/index given inputs and or transformations from other columns.
Implementation
Traditional index assignment. Advantages:
- Looks like a dictionary operation
- Overwrites the data frame
.assign() method. Advantage:
- It returns a dataframe so you can chain/pipe operations
- Can create multiple variables in a single call
- Easy to combine with numpy + lambda functions
- Improves readibility.
Let's see examples of both methods:
Transformation via index assignment¶
# With built in math operations
wc["av_goals_matches"] = wc["GoalsScored"]/wc["MatchesPlayed"]
wc.head()
## with numpy
wc["winner_and_hoster"] = np.where(wc.Country==wc.Winner, True, False)
wc[["Year", "Winner", "Country", "winner_and_hoster"]].head(5)
Transformation via apply methods by rows¶
As we saw before, apply() methods allow you to apply functions rowwise or columnwise in python. Here you are applying a certain data transformation to every row in your dataframe
# with an apply method + function
wc["av_goals_matches"] = wc.apply(lambda x: x["GoalsScored"]/x["MatchesPlayed"], axis=1)
wc.head()
.assign() method.¶
Allows you to create variables with methods chaining.
## when the winner was also hosting the world cup?
(
wc.
# multiple variables
assign(final = wc.Winner + " vs " + wc['Runners-Up'],
winner_and_hoster_np = np.where(wc.Winner==wc.Country, True, False),
av_goals_matches = lambda x: x["GoalsScored"]/x["MatchesPlayed"],
).
##allows for methods chaining
filter(["Year", "Winner", "Country", "final",
"winner_and_hoster_np", "av_goals_matches"]).
head(5))
Pay attention here! Using newly created variables!¶
.assign also allows for the use of newly create variables in the same chain. To do that, you need to make use of the lambda function
# Notice calling the recently created variable final
(wc.
assign(final = wc.Winner + " vs " + wc['Runners-Up'],
best_three = lambda x: x["final"] + " and " + x["Third"]).
filter(["best_three","final", "Country", "Third", "Runners-Up", "Winner"]).
head(5)
)
The combination of lambda (or a normal function) and .assign() is actually a nice property that resambles the properties of mutate in R. A lambda function (or a normal function) with .assign passes in the current state of the dataframe into the function. Because we create the variable in the state before, the lambda function allows us to access this new variable, and manipulate it in sequence. This also works for cases of grouped data, or chains in which we filter observations and transform the dataframe
To make this point clear, see how doing the same operation without a lambda function will throw an error:
# will throw an error
(wc.
assign(final= wc.Winner + " vs " + wc['Runners-Up'],
best_three = wc["final"] + " and " + wc["Third"]).
filter(["best_three","final", "Country"])
)
Renaming¶
Functionality:
Rename a columns directly or a via a function.
Implementation:
- Use dictionaries!
# Pandas: renaming variables using the rename method
wc.rename(columns={"Year":"year"}).head(3)
{col: col.lower() for col in wc.columns}
# we can use dictionary comprehension to apply functions in all
wc.rename(columns={col: col.lower() for col in wc.columns}).head(5)
# Or for only a set of the columns by given them as inputs
wc.rename(columns={col: col.lower() for col in ["Year", "Country"]}).head(5)
Practice¶
In this practice questions, we'll hone our data manipulation skills by examining conflict event data generated by the Armed Conflict Location & Event Data Project (ACLED). The aim is to practice some of the data manipulation functions covered in the lecture.
Data¶
ACLED is a "disaggregated data collection, analysis, and crisis mapping project. ACLED collects the dates, actors, locations, fatalities, and modalities of all reported political violence and protest events across Africa, South Asia, Southeast Asia, the Middle East, Central Asia and the Caucasus, Latin America and the Caribbean, and Southeastern and Eastern Europe and the Balkans." For this exercise, we'll focus just on the data pertaining to Africa. For more information regarding these data, please consult the ACLED methodology.
- Open the Data
- Create a new dataset with all columns with the words "event" or "actor"
- Rename all columns replacing the the word event to "event_acled_south_america"
Create three new columns:
- a binary representation for fatalities,
- a single column combining the columns: "region", "country",
- a new column recoding the inter1 variable with three string labels. These labels should indicate when the iter1 variable is equal to its median, below the median and above the median values.
# read
import pandas as pd
acled_sa = pd.read_csv("acled_south_america.csv")
acled_sa.columns
# 2
colnames = acled_sa.filter(like="event").columns.to_list() + acled_sa.filter(like="actor").columns.to_list()
acled_sa[colnames]
# another solution
acled_sa.filter(regex="event|actor").head(5)
# rename
acled_sa = acled_sa.rename(columns={col: col.replace("event", "event_acled_south_america") for col in acled_sa.columns}).head(5)
acled_sa
# 4
import numpy as np
(acled_sa.
assign(fatality_bin= np.where(acled_sa["fatalities"]>0, 1, 0),
full_location= acled_sa["region"]+ "-" + acled_sa["country"],
inter_recoded= np.where(acled_sa["inter1"]==acled_sa["inter1"].median(), "Median Value",
np.where(acled_sa["inter1"]>acled_sa["inter1"].median(),
"Higher Median", "Below Median"))
).head(5)
)
# solutions using np.select
median_value = acled_sa["inter1"].median()
# Define the conditions
conditions = [
acled_sa['inter1'] < median_value,
acled_sa['inter1'] == median_value,
acled_sa['inter1'] > median_value
]
# Define the output values
choices = ['below median', 'at median', 'above median']
# Use np.select
acled_sa['inter_recoded'] = np.select(conditions, choices, default='N/A')
# see
acled_sa.filter(["inter1", "inter_recoded"])
Row-Wise Operations¶
For data wrangling tasks at the rows of your data frame, we will discuss:
- Subsetting
- Filtering distinct values
- Recoding values
- Grouping and Summarizing
- Grouping and Transforming
- Sorting values
Subsetting¶
Functionality:
Slice the dataframe row-wise following a certain input.
Implementation:
- index based implementation
.locor.ilocmethods.query
Subsetting by index¶
# index based implementation
wc[wc.Year<1990]
# or with multiple condition
wc[(wc.Year<1990)&(wc.Year>1940)]
# see this
(wc.Year<1990)&(wc.Year>1940)
Subsetting with .loc() method¶
# pretty much the same + column names if you wish
wc.loc[(wc.Year<1990)&(wc.Year>1940),:]
Subsetting with .query(): a pipe approach¶
As before, you can use .query() methods to a more reable and pipeble approach. Notice the inside of the quotation marks use only the column name.
wc.query('Year<1990 & Year>1940')
Other types of subsetting¶
Subset by distinct entry¶
# Pandas: drop duplicative entries for a specific variable
# notice here you are actually deleting important rows
wc.drop_duplicates("Country")
Subset by sampling¶
# Pandas: randomly sample N number of rows from the data
wc.sample(10)
Recoding values¶
Functionality:
Recode a value given certain conditions. This type of transformation is one of the most importants in data cleaning.
Implementation:
There are many ways to recode variables in Python. We will showcase four of the most useful in my view.
First, we will see more generalized row-wise approach in pandas using:
map()apply()
Then we will see two vectorized solutions using numpy:
np.where()np.select()
Recode with map() + dictionaries¶
The map() function is used to substitute each value in a Series with another value.
- It takes a series as input
- Uses a function/dictionary to transform values
- Returns a series.
# map + dictionary to recode to create a dummy for certain country
# create a map function
mapping = {"Brazil":1}
# map the values
wc["brazil_winner"]= wc["Winner"].map(mapping)
# Fill missing values with a default value (e.g., 0)
wc["brazil_winner"].fillna(0, inplace=True)
# see results
wc.tail(5)
Recode with apply() + function¶
The apply() function is used when you want to apply a function along the axis of a DataFrame (either rows or columns). This function can be both an in-built function or a user-defined function.
# apply + function to recode
def get_dummies(x):
if x =="Brazil":
return 1
else:
return 0
# apply function
wc['Winner'].apply(get_dummies).head()
Notice, we can make it more general by providing a argument to the function
# apply + function to recode
def get_dummies(x, country):
if x == country:
return 1
else:
return 0
# Apply function with Uruguay now
wc["winner_uruguay"] = wc['Winner'].apply(get_dummies, country="Uruguay").head(10)
Here we are using apply for a column, so we do not need to indicate axis=1. That's also why we are inputing the columns as a serie. Notice we could do the same with assign:
# using apply inside of assign
wc.assign(winner_germany=wc['Winner'].apply(get_dummies, country="Germany"))
Summary of apply() and map()¶
apply()is used to apply a function along an axis of the DataFrame or on values of Series.- you are free to use lambda functions with map
- you can also apply functions column-wise changing the axis=1 argument.
map()is used to substitute each value in a Series with another value.
Recode using numpy¶
np.where: if-else approach.¶
np.whereis similar to ifelse in R- Useful if there’s only 1-2 (True/False conditions)
- sintax:
np.where(condition, true, false) - condition can be anything that returns a boolean.
Let's see some examples:
# create a new variable
wc["winner_brazil"]=np.where(wc["Winner"]=="Brazil", 1, 0)
wc.head()
# notice we can easily use np.where with assign
wc.assign(winner_brazil_=np.where(wc["Winner"]=="Brazil", 1, 0)).head(10)
# using string methods
(wc
.assign(south_america=np.where(wc["Winner"].str.contains("Brazil|Uruguay|Argentina"), 1, 0))
.filter(["Winner", "south_america"])
.head(10)
)
np.select: for multiple conditions¶
- np.select: similar to
case_whenin R. - useful for when there’s multiple conditions to be recoded
- sintax:
np.select(conditon, choicelist, default)
# step one: create a list of conditions
condition = [wc["Winner"]==wc["Country"],
wc["Runners-Up"]==wc["Country"],
wc["Third"]==wc["Country"]
]
# step two: create the choice list
choice_list = [1, 2, 3]
# recode
(wc
.assign(where_is_hoster=np.select(condition, choice_list, default="4+"))
.filter(["where_is_hoster"])
)
Group by: split-apply-combine¶
Functionality:
- Grouping data by specific variables/column indices
- Summarize/aggregate data by specific group features
How it works
“group by” is a common data wrangling process that exists in any language (R, Python, SQL) and refers to a process involving one or more of the following steps:
Splitting the data into groups based on some criteria.
Applying a function to each group independently.
Combining the results into a data structure.
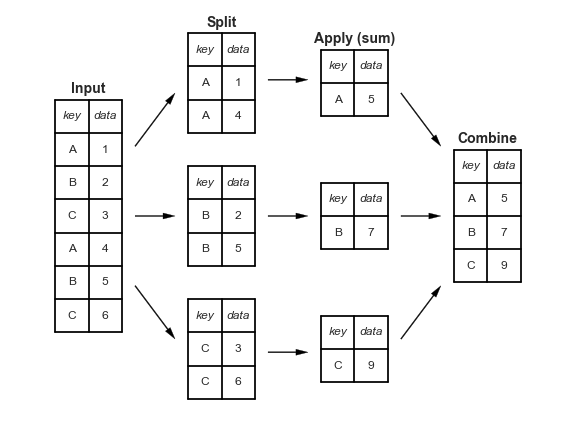
groupby (it just splits!)¶
The groupby() method in pandas splits your dataset in smaller parts. It generates an iterable where each group is broken up into a tuple (group,data).
# load worldcup dataset
import pandas as pd
wc = pd.read_csv("WorldCups.csv")
wc_matches = pd.read_csv("WorldCupMatches.csv")
#groupby object
g = wc.groupby(["Winner"])
g
The output of a groupby method is a flexible abstraction: although more complicated things are happening under the hood, in many ways, it can be treated as a collection of DataFrames. And as we learned, any collector can be iterated over!
# Iteration for groups
for group, data in g:
print(group)
# iteration for data grouped
for group, data in g:
print(data.head(2))
And we can acess a specific group:
g.get_group("Argentina")
Aggreations (summarize)¶
The power of a grouping function (like .groupby() shines when coupled with an aggregation operation.
An aggregation is any operation that reduces the dimensionality of the data!
pandas: .groupby() + built-in methods¶
# mean of all numeric variables grouping by winners
wc.groupby(["Winner"]).mean()
or select a specific variable to perform the aggregation step on.
# With a specific input
wc.groupby(["Winner"])["GoalsScored"].mean()
Notice the results come in a index structure. You can easily convert back to a fully formated dataframe by:
# reseting the index
wc.groupby(["Winner"])["GoalsScored"].mean().reset_index()
## as_index=false
wc.groupby(["Winner"], as_index=False)["GoalsScored"].mean()
Pandas offers many built-in methods to perform aggregations. Here is a list:
Built-in Methods¶
See user guide from Pandas Documentation
| Method | Functionality |
|---|---|
.any |
Compute whether any of the values in the groups are truthy |
.all |
Compute whether all of the values in the groups are truthy |
.count |
Compute the number of non-NA values in the groups |
.cov |
Compute the covariance of the groups |
.first |
Compute the first occurring value in each group |
.idxmax |
Compute the index of the maximum value in each group |
.idxmin |
Compute the index of the minimum value in each group |
.last |
Compute the last occurring value in each group |
.max |
Compute the maximum value in each group |
.mean |
Compute the mean of each group |
.median |
Compute the median of each group |
.min |
Compute the minimum value in each group |
.nunique |
Compute the number of unique values in each group |
.prod |
Compute the product of the values in each group |
.quantile |
Compute a given quantile of the values in each group |
.sem |
Compute the standard error of the mean of the values in each group |
.size |
Compute the number of values in each group |
.skew |
Compute the skew of the values in each group |
.std |
Compute the standard deviation of the values in each group |
.sum |
Compute the sum of the values in each group |
.var |
Compute the variance of the values in each group |
Let's see some examples:
# max goal by team as home team
(wc_matches
.groupby(["Home Team Name"])
["Home Team Goals"]
.max()
.reset_index()
.sort_values("Home Team Goals", ascending=False))
# number of matches as home team
(wc_matches
.groupby(["Home Team Name"], as_index=False)
["Home Team Name"]
.size()
.sort_values("size", ascending=False)
)
pandas: .groupby() + agg()¶
Alternatively, we can specify a whole range of operations to aggregate by (along with specific variable columns) using the .aggregate()/.agg() method. To keep track of which operations correspond which variable, pandas will generate a hierarchical index for column entries.
wc.groupby(["Winner"])["GoalsScored"].agg(["mean","std","median"]).reset_index()
We can also user-defined functions into the aggregate() function as well.
import numpy as np
def mean_add_50(x):
return np.mean(x) + 50
wc.groupby(["Winner"])["GoalsScored"].agg(["mean","std","median",mean_add_50])
agg() + .rename() provides an easy workflow to rename your newly created variables
(wc.groupby(["Winner"])["GoalsScored"].
agg(["mean","std","median",mean_add_50]).
rename(columns={"mean": "goals_mean",
"std": "goals_std",
"median": "goals_median",
"mean_add_50":"mean_50_goals"})
)
pandas: .groupby() + .apply()¶
Even though I cover this in more details on the miscellaneous notebook, here is a good moment to bring the .apply() methods from pandas one more time.
The apply method lets you apply an arbitrary function by row or by columns on your data frame. As you can imagine, it also allow you to apply results by at a groupby object, and returns a summarizes dataset.
The function should take a DataFrame and returns either a Pandas object (e.g., DataFrame, Series) or a scalar
Let's an example with the mean add function:
(wc.groupby(["Country"])["GoalsScored"].
apply(mean_add_50).
reset_index()
)
multi-index grouping¶
We can also group by more than one variable (i.e. implement a multi-index on the rows).
wc.groupby(["Winner", "Year"])["GoalsScored"].mean().reset_index().head(5)
pandas: .groupby() + .transform()¶
Other times we want to implement data manipulations by some grouping variable but retain structure of the original data. Put differently, our aim is not to aggregate but to perform some operation across specific groups.
To do so, we will combine .groupby() and .transform() methods. Let's see an example:
# create a new column
# notive you need to select the column you want to transform.
wc.groupby(["Winner"])["GoalsScored"].transform("mean")
# easily combined with assign
# create a new column
(wc.assign(goals_score_wc_mean_wc=wc.groupby(["Winner"])["GoalsScored"].
transform("mean")))
# also: very useful with lambda functions
wc.groupby("Winner")["GoalsScored"].transform(lambda x: x - x.mean())
Sorting values¶
# Pandas: sort values by a column variable (ascending)
wc.sort_values('Country').head(3)
# Pandas: sort values by a column variable (descending)
wc.sort_values('Year',ascending=False).head(3)
# Pandas: sort values by more than one column variable
wc.sort_values(['Winner', "Country"]).head(3)
That was a lot! but you will get to keep this notebook for you!¶
And remember of the cheat sheet for pandas
Practice¶
Using the same ACLED data from the previous exercise, answer:
What are the different event types recorded?
How many events are recorded for each year?
What’s the most common event type in the data?
Which countries had the highest number of reported fatalities?
# response
!jupyter nbconvert _week-6c-data_wrangling_pandas.ipynb --to html --template classic